Estimation
To start with, I will use the RandomForest estimator and see how it does.
clf = RandomForestClassifier(n_estimators=100, max_depth=5, random_state=1)
clf.fit(x_train, y_train)
y_pred = clf.predict(test_df)
# score = 0.78468
Class rebalancing
However, our data is actually imbalanced with more people dying than surviving (Survived feature has more 0s than 1s).
y_train.value_counts()
# Output
# 0.0 549
# 1.0 342
So let's do some resampling with SMOTE.
from imblearn.over_sampling import SMOTE
resampler = SMOTE(random_state=1)
x_train_resample, y_train = resampler.fit_resample(x_train, y_train)
x_train = pd.DataFrame(x_train_resample, columns=x_train.columns)
Hyperparameter Tuning
It is also important to find the best hyperparameters for the estimator. For this, I will use Optuna, which is not only much faster than GridSearch but can also scan a much wider range of values.
To find the most optimal hyperparameters with Optuna, make a function that creates the estimator with variable hyperparameters and return the cross validation score of that estimator. Then make a study and run a number of 'trials' (iterations) to optimize the score returned by the function (either maximize or minimize).
import optuna
def objective(trial):
params = {'n_estimators': trial.suggest_int('n_estimators', 10, 500),
'max_depth': trial.suggest_int('max_depth', 2, 12),
'criterion': trial.suggest_categorical('criterion', ['gini', 'entropy']),
'min_samples_split': trial.suggest_int('min_samples_split', 2, 4),
'min_samples_leaf': trial.suggest_int('min_samples_leaf', 1, 4),
'ccp_alpha': trial.suggest_float('ccp_alpha', 0, 0.5)}
clf = RandomForestClassifier(**params, random_state=1)
score = cross_val_score(clf, x_train, y_train)
return score.mean()
study = optuna.create_study(direction="maximize")
study.optimize(objective, n_trials=100)
study.best_trial
# n_estimators: 192, max_depth: 7, criterion: 'gini', min_samples_split: 4, min_samples_leaf: 4, ccp_alpha: 0.00010049065148174871
rf1 = RandomForestClassifier(n_estimators=192, max_depth=7, criterion='gini', min_samples_split=4, min_samples_leaf=4, ccp_alpha=0.00010049065148174871, random_state=1)
rf1.fit(x_train, y_train)
y_pred = rf1.predict(test_df)
# score = 0.78947
Balancing classes and tuning hyperparameters increases accuracy to 0.78947.
After trying a umber of estimators, I finally stacked RandomForest, ExtraTrees and XGBoost together and tuned hyperparameters:
from sklearn.ensemble import StackingClassifier
def objective(trial):
xgb_params = {'n_estimators': trial.suggest_int('xgb_n_estimators', 10, 500),
'max_depth': trial.suggest_int('xgb_max_depth', 2, 12),
'learning_rate': trial.suggest_float('learning_rate', 0, 1),
'colsample_bytree': trial.suggest_float('colsample_bytree', 0, 1),
'colsample_bylevel': trial.suggest_float('colsample_bylevel', 0, 1),
'colsample_bynode': trial.suggest_float('colsample_bynode', 0, 1)}
rf_params = {'n_estimators': trial.suggest_int('rf_n_estimators', 10, 500),
'max_depth': trial.suggest_int('rf_max_depth', 2, 12),
'criterion': trial.suggest_categorical('rf_criterion', ['gini', 'entropy']),
'min_samples_split': trial.suggest_int('rf_min_samples_split', 2, 4),
'min_samples_leaf': trial.suggest_int('rf_min_samples_leaf', 1, 4),
'ccp_alpha': trial.suggest_float('rf_ccp_alpha', 0, 0.5)}
et_params = {'n_estimators': trial.suggest_int('et_n_estimators', 10, 500),
'max_depth': trial.suggest_int('et_max_depth', 2, 12),
'criterion': trial.suggest_categorical('et_criterion', ['gini', 'entropy']),
'min_samples_split': trial.suggest_int('et_min_samples_split', 2, 4),
'min_samples_leaf': trial.suggest_int('et_min_samples_leaf', 1, 4),
'ccp_alpha': trial.suggest_float('et_ccp_alpha', 0, 0.5)}
passthrough = trial.suggest_categorical('passthrough', [True, False])
xgb2 = XGBClassifier(**xgb_params, random_state=1)
rf2 = RandomForestClassifier(**rf_params, random_state=1)
et2 = ExtraTreesClassifier(**et_params, random_state=1)
clf = StackingClassifier(estimators=[('et2', et2), ('xgb2', xgb2)], final_estimator=rf2, passthrough=passthrough)
score = cross_val_score(clf, x_train, y_train)
return score.mean()
study = optuna.create_study(direction="maximize")
study.optimize(objective, n_trials=100)
study.best_trial
# xgb_n_estimators: 224, xgb_max_depth: 9, learning_rate: 0.6678429954436355, colsample_bytree: 0.45970974209639903,
# colsample_bylevel: 0.6689249222061762, colsample_bynode: 0.0731575107142313, rf_n_estimators: 424, rf_max_depth: 10,
# rf_criterion: 'entropy', rf_min_samples_split: 3, rf_min_samples_leaf: 4, rf_ccp_alpha: 0.0010533704079154983,
# et_n_estimators: 485, et_max_depth: 4, et_criterion: 'gini', et_min_samples_split: 2, et_min_samples_leaf: 4,
# et_ccp_alpha: 0.05935569112508277, passthrough: True
xgb2 = XGBClassifier(n_estimators=224, max_depth=9, learning_rate=0.6678429954436355, colsample_bytree=0.45970974209639903, colsample_bylevel=0.6689249222061762, colsample_bynode=0.0731575107142313, random_state=1)
rf2 = RandomForestClassifier(n_estimators=424, max_depth=10, criterion='entropy', min_samples_split=3, min_samples_leaf=4, ccp_alpha=0.0010533704079154983, random_state=1)
et2 = ExtraTreesClassifier(n_estimators=485, max_depth=4, criterion='gini', min_samples_split=2, min_samples_leaf=4, ccp_alpha=0.05935569112508277, random_state=1)
clf = StackingClassifier(estimators=[('et2', et2), ('xgb2', xgb2)], final_estimator=rf2, passthrough=True)
clf.fit(x_train, y_train)
y_pred = clf.predict(test_df)
# score = 0.78947
Stacking with completely different hyperparameters gives the exact same score as just RandomForest. They definitely don't make the exact same predictions since their prediction value counts are different, but somehow stacking resulted in correctly classifying exactly as many samples that RandomForest had misclassified, as it misclassified samples that RandomForest had correctly classified.
Model Explanation
Let's try to understand what my model is doing. First find the Permutation Importance score of features and then make a SHAP plot. However, since SHAP doesn't support StackingClassifier yet, I will be doing Permutation Importance and SHAP analysis of the individual RandomForest that got the joint highest accuracy instead.
perm = PermutationImportance(rf1, random_state=1).fit(x_train, y_train)
eli5.show_weights(perm, feature_names = x_train.columns.tolist())
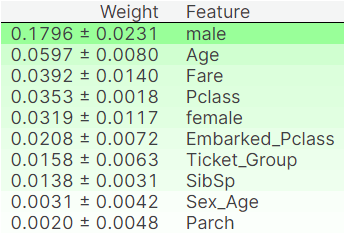
The gender, age and fare of a passenger seems to affect prediction the most. This makes sense - females, children and the affluent (those who have bought expensive ticket) are usually given priority in such situations.
The number of parents and children of a passenger seems to affect prediction the least.
plot, score = shap_score(rf1, x_train)
plot
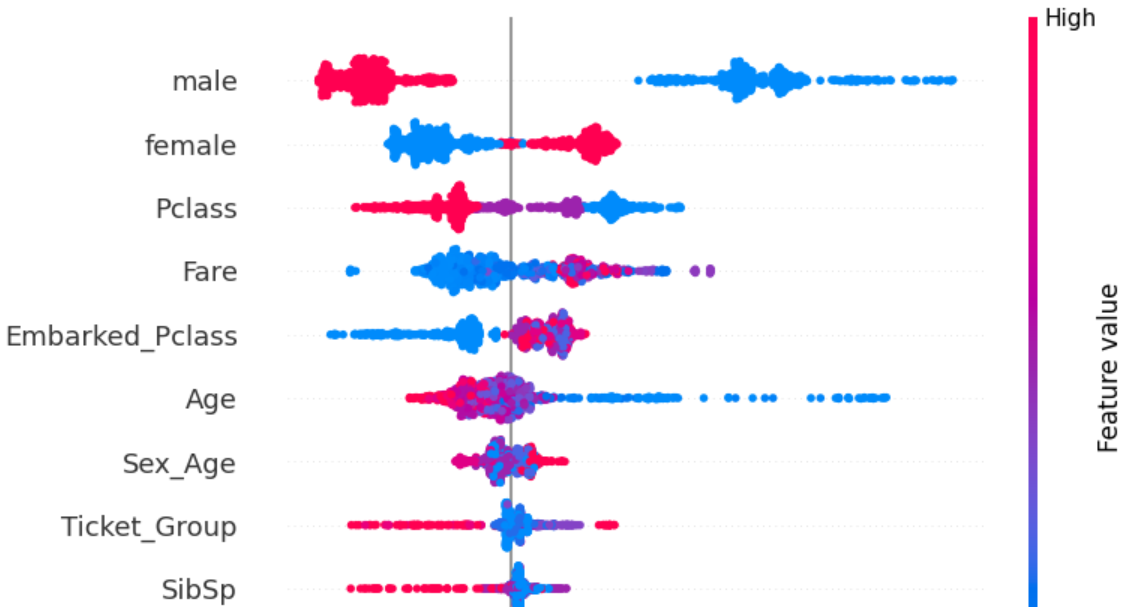
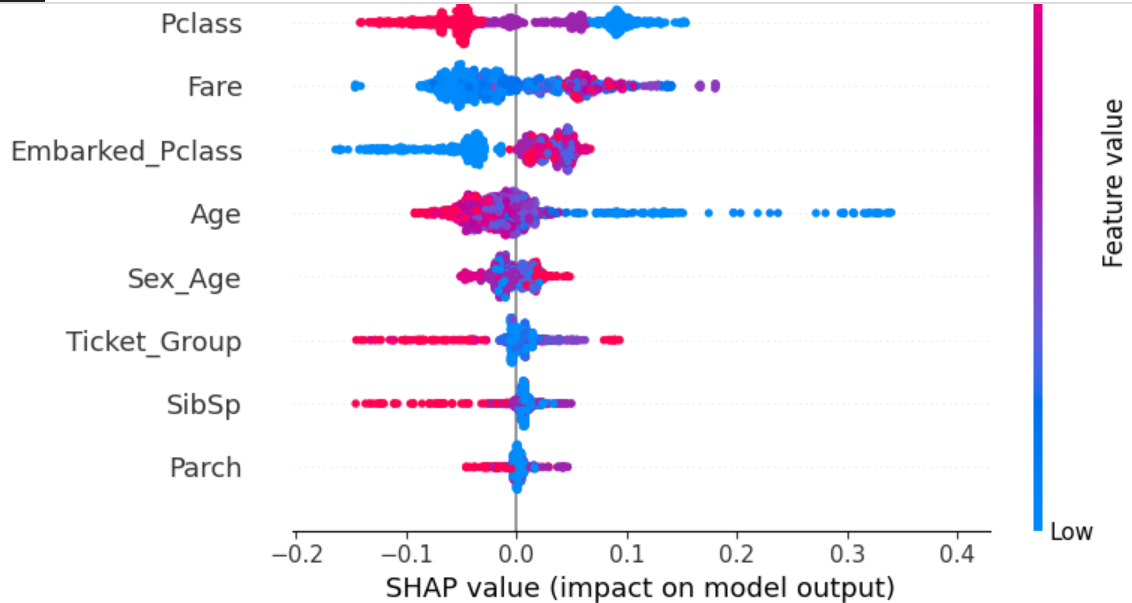
The SHAP plot gives more comprehensive understanding.
Those having Pclass 3 have less survivability while those having Pclass 1 have higher survivability. This directly corresponds with the fact that those with lower Fare have lower survivability than those with higher Fare.
Passengers with lower Age (children) have significantly higher survivability than older passengers.
Ticket_Group is interesting. Those who bought tickets in large groups have lower survivability. Perhaps this is because only poorer people would be more likely to buy ticket in a group. This needs more investigation.
Let's take an arbitrary sample from the test set and see how different features affect its prediction.
explainer = shap.TreeExplainer(rf1)
shap_values = explainer.shap_values(test_df.iloc[0])
shap.initjs()
shap.force_plot(explainer.expected_value[1], shap_values[1], test_df.iloc[0])
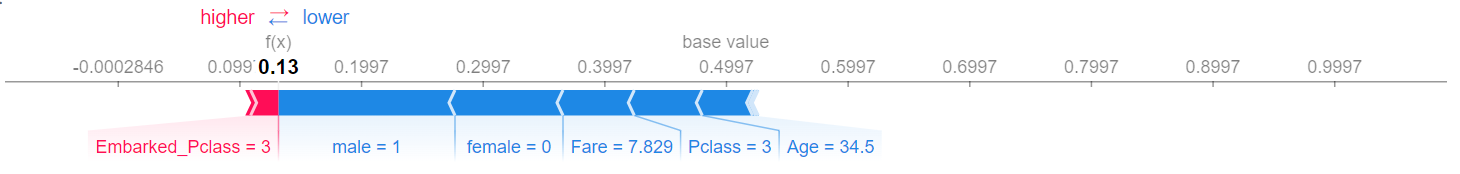
For this passenger, prediction is most affected by him being a male and not a female, that he has bought ticket worth only 7.829, that he is a class 3 passenger and he is 34.5 years old (all of which decreases survivability a lot), as well as that he belongs to Embarked_Pclass cluster 3 which increases survivability slightly. In total, he gets a survivability of only 13%.